At initiation DnaA-ATP molecules cooperatively bind the left half of the origin to form a right-handed DnaA-ATP helix, where individual DnaA molecules interact through their AAA+ domains, with oriC DNA wrapped around it. Binding of IHF immediately upstream of the DUE flanking R1 DnaA-box introduces a 160u bend in the DNA reversing the orientation of the DNA helical axis and assist in melting the DUE region. One of the exposed single-stranded DUE regions is fixed by binding the existing DnaA-ATP helix while the other strand is exposed for DnaC assisted DnaB helicase loading by the DnaA molecule bound to the R1 box. Further opening of the duplex allows for loading of the second helicase by one or more N-terminal domains of the DnaA-ATP filament. Although SJN 2511 promoted by formation of a DnaA oligomer on oriC, the exact mechanism for helicase loading at the origin differ between bacteria. After helicase loading, a cascade of events leading to replisome assembly and the beginning of the elongation follows. The replisome structure was recently covered in an excellent review and consists of a primosome complex and a PolIII holoenzyme complex, where each PolIII holoenzyme complex can be further divided into three different complexes: PolIII core, the sliding clamp and the clamp loader. The core polymerase needs the sliding clamp for processivity, which in turn is loaded onto the DNA by the clamp loader. In the firmicutes including S. aureus, the process of elongation is similar to that in E. coli with a couple of notable exceptions. The S. aureus helicase is loaded by the DnaI helicase loader assisted by the DnaB and DnaD proteins and two different replicative polymerases are used. The DnaE which is homologous to the E. coli PolIIIa only extends RNA AZ 960 primers initially and hands them off to PolC which is responsible for the processive synthesis. A third difference was recently revealed. Primer hand off in Bacillus subtilis, can occur after the synthesis of only two nucleotides by the DnaG primase and does not require other replication proteins. This is in contrast to the three-point switch hand off mechanism in E. coli. Here the x polypeptide of the clamp loader interacts with SSB to displace DnaG from the SSB-DnaG complex resulting in release of the primer which is then extended by the processive polymerase. In all bacteria examined so far the ring shaped b-clamp is a homodimer which encircles the DNA and slides along the duplex bringing the polymerase into contact with the DNA to ensure processivity. The b-clamp interacts with many different proteins including DnaE, PolC, d, PolIV, PolV, PolI, MutS, MutL, DNA ligase and Hda. These proteins all contain a conserved b-binding motif which binds a hydrophobic pocket located in each DnaN protomer. The b-sliding clamp has been the target 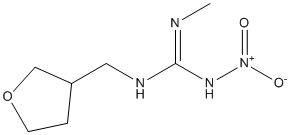 for potential new antibiotics and two different approaches have been used to identify compounds that block the peptide-binding pocket of b. First, synthetic peptides containing the beta-binding domain QLD/SLF were found to inhibit PolC-b2 and d-b2 interactions and similarly peptides containing b-binding sequence from d and Hda bound the b-clamp and inhibited DNA synthesis in vitro. Subsequently more efficient binders were identified by modification of the b-binding domain and these optimized peptide motifs have served as starting point for small molecule mimics to identify compounds that inhibit the a-b2 interaction at micromolar concentrations. In the second approach, a fluorescence based peptide displacement assay was used to identify small compounds that bind to the peptide-binding pocket of b. One compound, RU7, which inhibited PolII, PolIII and PolIV although to different extents was identified from a collection of 30,600 polar organic compounds. It was suggested that RU7 can be used as a starting point for rational drug design to create stronger inhibitors of replication. A fairly unexploited class of compounds that has attracted attention as putative antimicrobials is peptides. The extensively studied natural antimicrobial peptides are produced by multicellular organisms and the majority act by insertion and alteration/ damage of cytoplasmic membranes via formation of ion channels or transmembrane pores, but other have been associated with intracellular targets such as DNA and RNA synthesis and inhibition of enzymatic activities. This indicates that certain peptides can traverse the bacterial membrane to find their intracellular targets.
for potential new antibiotics and two different approaches have been used to identify compounds that block the peptide-binding pocket of b. First, synthetic peptides containing the beta-binding domain QLD/SLF were found to inhibit PolC-b2 and d-b2 interactions and similarly peptides containing b-binding sequence from d and Hda bound the b-clamp and inhibited DNA synthesis in vitro. Subsequently more efficient binders were identified by modification of the b-binding domain and these optimized peptide motifs have served as starting point for small molecule mimics to identify compounds that inhibit the a-b2 interaction at micromolar concentrations. In the second approach, a fluorescence based peptide displacement assay was used to identify small compounds that bind to the peptide-binding pocket of b. One compound, RU7, which inhibited PolII, PolIII and PolIV although to different extents was identified from a collection of 30,600 polar organic compounds. It was suggested that RU7 can be used as a starting point for rational drug design to create stronger inhibitors of replication. A fairly unexploited class of compounds that has attracted attention as putative antimicrobials is peptides. The extensively studied natural antimicrobial peptides are produced by multicellular organisms and the majority act by insertion and alteration/ damage of cytoplasmic membranes via formation of ion channels or transmembrane pores, but other have been associated with intracellular targets such as DNA and RNA synthesis and inhibition of enzymatic activities. This indicates that certain peptides can traverse the bacterial membrane to find their intracellular targets.