Most in vitro models of Mtb dormancy are able to induce non-replicating state in mycobacteria but they have not been able to demonstrate all the above characteristics of dormant mycobacteria. Recently, Deb et. al. developed a multiple stress model of Mtb dormancy in an attempt to replicate the conditions the pathogen is thought to encounter in host granuloma, and showed that Mtb indeed exhibited dormancy as characterized by all the hallmark characteristics of dormancy, however this model does not involve host-pathogen interaction. In lipid loaded THP-1 derived macrophages Mtb has been found to go into a dormant state. At low dose infection, which closely mimics the in vivo scenario, Mtb in our granuloma model displays characteristics of dormant Mtb, via. development of Rif-tolerance, loss of acid fastness and accumulation of lipids bodies. A SILAC approach was designed to identify pathways associated with bladder cancer aggressiveness. Cul3 was revealed as a candidate contributing to the aggressive phenotype of T24T modifying cytoskeleton remodelling and as a bladder cancer biomarker correlating with poor outcome. Our comparative functional analyses of T24-T24T were complementary and agreed with previous in vitro results describing a more aggressive phenotype of T24T cells. By contrast to earlier analyses, we performed proliferation by seeding cells at a three-fold higher density, plus wound healing and invasion assays. These data highlighted the ability of T24T cells to grow on top of each other, in contrast to the contact inhibition previously described for T24 cells. These results further suggested that T24T cells have a greater Folinic acid calcium salt pentahydrate potential for proliferation, motility and potentially to metastasize, as demonstrated in vivo. A high number of proteins were found differentially expressed between T24-T24T, with biological network annotations supporting the functional differences observed in vitro. Furthermore, proteins were shown differentially expressed using oligonucleotide arrays and by selected immunoblotting. Immunostaining of tissue arrays containing independent series of bladder cancer patients served to assess the associations of a selected 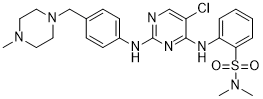 protein, Cul3, with clinicopathological variables. Functional analyses and immunoblotting validation upon Cul3 silencing highlighted its impact in the aggressive phenotype of T24T cells and at modulating other cytoskeleton proteins identified by SILAC. Thus, combination of -omic approaches, functional and clinical analyses identified Cul3 as a novel candidate related to bladder cancer aggressiveness. The extent of the proteomic profile defined in this study was comparable to other SILAC studies. On the basis of the identity and biological abundance of the proteins identified, SILAC exhibited a satisfactory dynamic range in profiling both high- and low-abundance proteins. The broad spectrum of proteins observed reflects SILAC suitability for proteomic studies of cancer cells. Subcellular Mepiroxol fractionation reduced sample complexity and increased the probability of detecting less abundant proteins. The level of ambiguity for a protein ratio was estimated taking into account the SDs within each protein because every SILAC ratio was calculated as a mean of at least 2 peptide values with their associated SDs. We selected 1.5 and 0.67 as cutoffs, also frequently used in SILAC-related studies. When comparing two closely-related cell lines, it is expected that most of the proteins are expressed at similar levels. Indeed, most of the SILAC ratios were within the 0.67�C1.5 range.
protein, Cul3, with clinicopathological variables. Functional analyses and immunoblotting validation upon Cul3 silencing highlighted its impact in the aggressive phenotype of T24T cells and at modulating other cytoskeleton proteins identified by SILAC. Thus, combination of -omic approaches, functional and clinical analyses identified Cul3 as a novel candidate related to bladder cancer aggressiveness. The extent of the proteomic profile defined in this study was comparable to other SILAC studies. On the basis of the identity and biological abundance of the proteins identified, SILAC exhibited a satisfactory dynamic range in profiling both high- and low-abundance proteins. The broad spectrum of proteins observed reflects SILAC suitability for proteomic studies of cancer cells. Subcellular Mepiroxol fractionation reduced sample complexity and increased the probability of detecting less abundant proteins. The level of ambiguity for a protein ratio was estimated taking into account the SDs within each protein because every SILAC ratio was calculated as a mean of at least 2 peptide values with their associated SDs. We selected 1.5 and 0.67 as cutoffs, also frequently used in SILAC-related studies. When comparing two closely-related cell lines, it is expected that most of the proteins are expressed at similar levels. Indeed, most of the SILAC ratios were within the 0.67�C1.5 range.